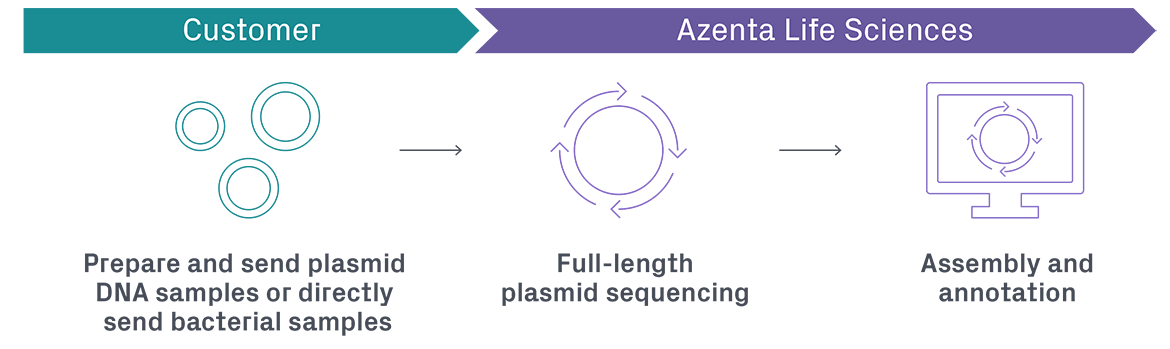
In the current sequencing landscape, there are two major technologies that are often used to read the nucleotide bases in plasmid DNA: the traditional Sanger method and long-read sequencing using Oxford Nanopore Technology (ONT). These two methods have a few fundamental differences and work best when used synergistically for an accurate and comprehensive view into the complete plasmid sequence.
In this blog, explore the cutting-edge realm of genomic research with Oxford Nanopore Sequencing (ONT), and how this innovative NGS technology offers real-time, long-read capabilities, revolutionizing our approach to decoding genetic information.
What is ONT?
Oxford Nanopore Sequencing, or ONT, is a third-generation sequencer that generates high-quality long reads.
How?
It generates these leads with amplification-free library prep. Unlike PCR-based amplification, which can introduce unwanted artifacts, the DNA strand is separated, enters the nanopore protein, and an electric current with Oxford Nanopore technology will run through the single-stranded DNA, measuring the nucleotides in real time.
Once you are satisfied with the amount of data that is generated, you can stop the run. Results are generated and uploaded rapidly, and there is no set limit on cycles.
How does ONT differ from Sanger Sequencing?
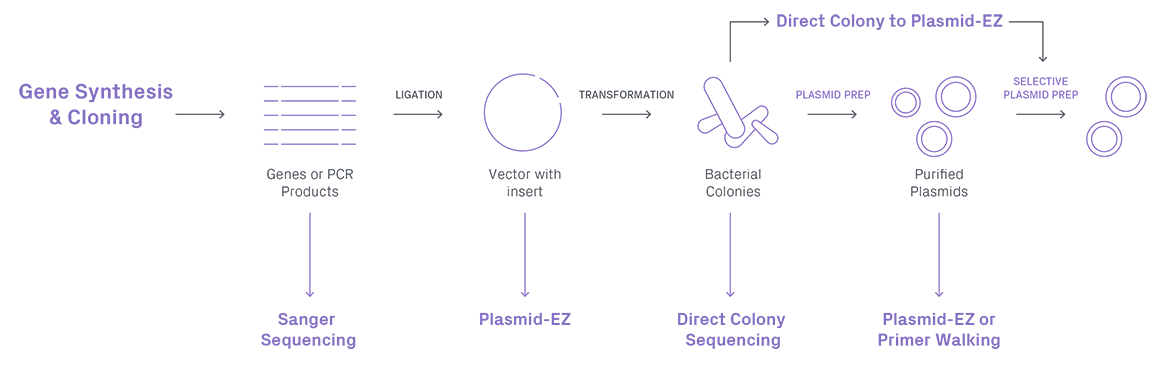
Sanger sequencing is particularly well-suited for plasmid sequence validation due to its accuracy in reading short DNA fragments. Therefore, it is a reliable method to focus on specific regions of the plasmids, such as gene insert sequences that need to be validated.
Unlike Sanger sequencing, the Oxford Nanopore platform excels in generating exceptionally long reads, making it easier to capture complete plasmid sequences with a few rounds of sequencing. This capability is invaluable when dealing with complex plasmid structures, allowing for a comprehensive understanding of gene arrangements, regulatory elements, and other critical features.
Sample preparation for plasmid sequencing with ONT is streamlined and does not require primers, eliminating the need for labor-intensive steps associated with traditional methods.
While ONT offers strong advantages in read length and real-time monitoring, there are a few potential challenges such as higher error rates, particularly in homo polymeric regions, that make it less than ideal for sequence confirmation with complete accuracy. Thus, it is recommended to leverage both ONT and Sanger together for optimal results.
| Sanger Sequencing | Oxford Nanopore Technology (ONT) | |
|---|---|---|
| Read Length | Shorter reads, typically less than 1000 bp | Longer reads, typically 10kb or higher |
| Accuracy | High accuracy, low error rate | Higher error rates, particularly in homopolymeric regions |
| Throughput and Cost | Lower throughput, higher cost per base | Higher throughput, lower cost per base with high sample number |
| Sample Preparation | Tedious and time-consuming sample preparation involving cloning and amplification | Streamlined sample preparation, reducing time and complexity of workflows |
| Primers | Requires primer design to amplify regions of interest | No primer design necessary, entire plasmid can be sequenced at once |
| Suitability for Plasmids | Precise and reliable for short sequences; well-suited for validation of specific regions within the plasmids | Suitable for generating full plasmid sequence and annotation maps |
Why is ONT used for plasmid sequencing with Plasmid-EZ?
ONT is used for Plasmid-EZ because it offers better efficiency at a lower cost, and with more convenience.
Compared to Sanger sequencing which offers shorter reads (typically less than 1000 bp), ONT provides a far more efficient way to sequence with longer reads (typically 10kb or higher), higher throughput, and lower cost per base. Our Plasmid-EZ service offers a rapid turnaround time by utilizing ONT to streamline sample preparations and rapidly sequence whole plasmids, contributing to a complete end-to-end workflow. Sanger is great for validating short sequences of specific regions due to its high accuracy, but if you’re looking to generate full plasmid sequences and annotation maps then ONT is the tool of choice.
What does a sample submission look like?
Visit our Sample Submission Guidelines for info on the size, minimum amount, concentration, purity, and buffer for best sequencing results.
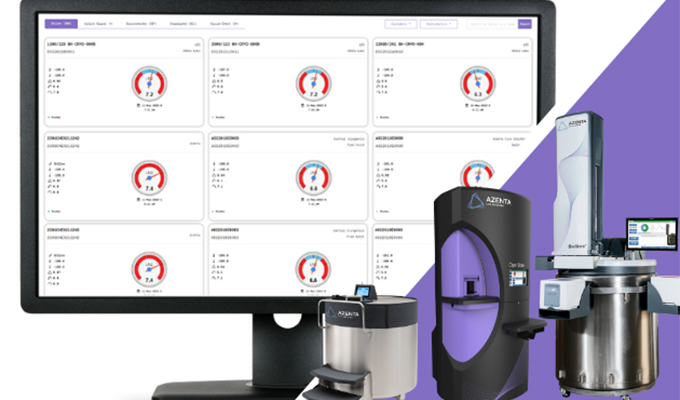
What does our sequencing workflow look like?
Send us your sample, and we’ll take care of the rest.
Our deliverables include:
- FASTQ files (raw data from ONT)
- Read-length and quality report
- Consensus plasmid assembly
- Plasmid annotation files
What are some limitations of ONT with Plasmid-EZ?
One limitation is that the sample must meet a 50 ng/μL concentration, as a low concentration causes an increase in fragmentation and a low number of reads.
Poly A and homopolymer regions may also be truncated in ONT and would benefit from follow-up Sanger sequencing for that specific region.
Lastly, ONT is set up for clonal populations, rather than sequencing multiple plasmids in a mixed sample.
For analyzing mutagenesis, would Sanger or ONT work best?
If you are growing and isolating individual colonies and looking at several mutations throughout the entire plasmid, Plasmid-EZ with ONT is best.
If you are creating and analyzing a small mutation, such as 500 base pairs, Sanger is recommended.
For random mutants that have a plasmid containing a mixture of different mutations (not growing out colonies), a variant analysis with NGS is advised.
How are Sanger and ONT successfully used together?
While ONT offers strong advantages in read length and real-time monitoring, there are a few potential challenges such as higher error rates, particularly in homopolymeric regions, that make it less than ideal for sequence confirmation with complete accuracy.
That’s where Sanger sequencing comes in— since Sanger has higher read accuracy for short DNA fragments, it is particularly well-suited for validating challenging regions of a plasmid sequence, such as gene insert sequences.
By knowing some of the limitations of your plasmid sample and the capabilities of ONT, you can attain the best turnaround time and lowest cost by using a combination of the two services.
Our Technical Support team can provide recommendations for your custom research needs and help align both whole plasmid sequencing data and Sanger data on the same construct.
Integrate ONT Seamlessly with Plasmid-EZ
Oxford Nanopore Sequencing within our Plasmid-EZ service brings forth efficiency, cost-effectiveness, and a streamlined workflow for genomic analysis. Its rapid turnaround time and end-to-end capabilities make it a tool that seamlessly integrates into your procedure, offering a practical and advanced solution for your sequencing needs. Experience the power of ONT within our service and propel your genomic research forward with confidence.
About the Authors

Annie Huang
Annie Huang is a Marketing Manager for Genomics content strategy at Azenta Life Sciences. She holds a BS in Microbiology and Molecular Genetics from Michigan State University and has worked in Product Marketing, Technical Sales, and Market Development across the life sciences industry prior to her current role. Today, she oversees content management and strategy for Next Generation Sequencing, Synthetic Biology (Oligo, Gene Synthesis, Plasmid Prep), and PCR + Sanger Sequencing portfolios for GENEWIZ Multiomics & Synthesis Solutions from Azenta Life Sciences.

Bhagyashree Birla
Bhagyashree Birla is a Senior Technical Specialist for GENEWIZ Multiomics & Synthesis Solutions from Azenta Life Sciences. She earned her PhD from Iowa State University and has held technical scientist roles at various life science organizations. In her current position, she advises researchers on next generation sequencing project design and best approaches to set them up for success.