Tools for Simplifying Single-Cell Data Analysis
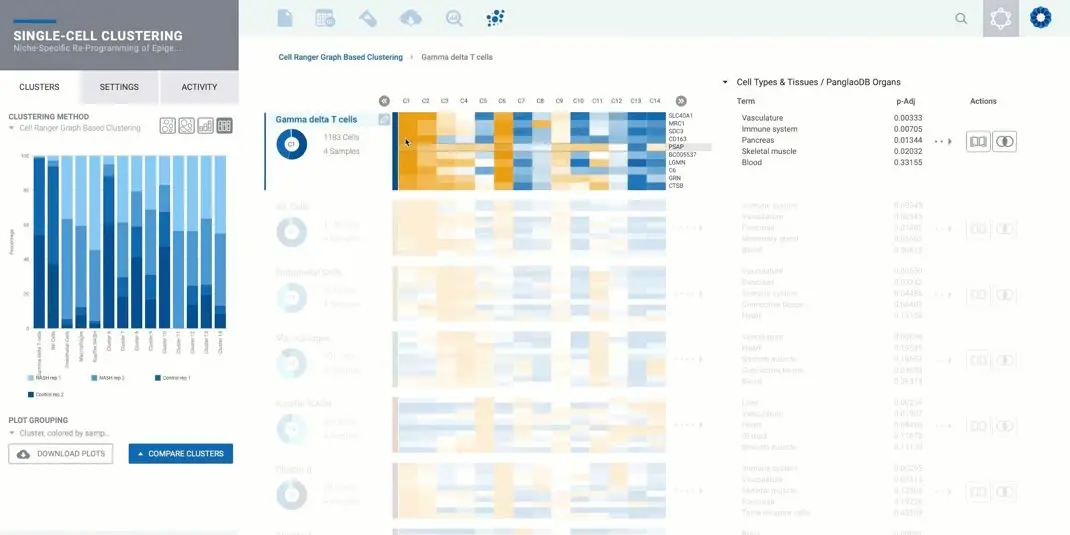
Examining gene expression at single-cell resolution provides insights into complex populations by identifying rare cell types, revealing cell heterogeneity, and uncovering other unique gene expression signatures that can guide our understanding of disease – and ultimately – drug development. However, finding meaningful patterns in single-cell RNA sequencing (scRNA-Seq) results can be challenging, as data sets generated from these experiments are often very large and complex.
To help researchers gain these insights, the Next Generation Sequencing (NGS) team at GENEWIZ has partnered with ROSALIND to offer a complete sequencing to data analysis approach for analyzing scRNA-Seq data. Leveraging GENEWIZ’s tried-and-true scRNA-Seq service and ROSALIND’s intuitive data analysis portal, this powerful workflow enables researchers to quickly obtain deep insights from their data, including interpretation with gene set enrichment and variation analysis, automated gene clustering, pathway interpretation, and publication-quality figures. We recently sat down with Jean Lozach, ROSALIND’s chief scientific officer, to learn more about the power of this data portal and their partnership with GENEWIZ.
Coming from an extensive background in NGS and bioinformatics, what was most interesting to you about joining ROSALIND and its platform for data analysis?
Jean Lozach (JL): My goal when I joined ROSALIND (called OnRamp Bio at the time) was to develop an analysis/bioinformatics platform for biologists. After 15 years in the genomics space and the fast rate of adoption of NGS, I realized that the shortage of bioinformaticians was a real problem affecting researchers everywhere. From academia to biotech, and even in biopharma, scientists must wait to have their data analyzed.
In early 2017, we decided to build a new platform to give any researcher the ability to analyze and interpret their experiments in just a few hours after obtaining raw data files. This was really the driver for me joining the team. I wanted us to build a platform that was not only scientifically sound but also had an intuitive user interface that biologists could use without having to learn how to run command line scripts. I think our team really achieved this goal at ROSALIND.
What is most unique about ROSALIND’s single-cell data analysis platform?
JL: Two features of the experience are truly unique. The first is how we analyze the data: each original sample is split into many smaller samples based on the cell types identified, allowing us to provide the same experience as bulk RNA-Seq. This is extremely powerful as each scRNA-Seq comparison can be combined with results from bulk RNA-Seq, NanoString® gene expression panels, or even ChIP-Seq to build a multi-omics, meta-analysis approach to identify patterns in your data.
The second unique feature is an intelligent system to annotate the identified clusters. It’s always a challenge to identify the cell type of each cluster, but the platform is helping researchers by suggesting a few potential cell types for each cluster based on the markers identified in the cluster. Overall, the platform offers a very simple way to go from the FASTQ files generated from your 10x Genomics® scRNA-Seq experiment to biological interpretation in just a few hours, without the need to access a server or run any command line instruction.
One concern could be data security. Can you share a bit more on cybersecurity?
JL: Security is often a concern, as it should be. The platform is built on the Google® Cloud infrastructure inside a virtual private cloud. Security measures are in place to ensure that all data is safely encrypted at rest and during transfer. We are audited on a regular basis by biopharma and biotech companies and have ALWAYS passed all their tests successfully. The platform is even compliant with FedRAMP (Federal Risk and Authorization Management Program), which is the mandatory cyber security risk management program required by the government when working with US federal agencies. The platform was used extensively by the NIH and FDA during the COVID-19 pandemic, for example. Of course, we can provide documents explaining in more detail all the security systems and features of the platform to ensure that our customers’ data is and always stays safe.
What excites you about ROSALIND partnering with GENEWIZ?
JL: I’m really excited about this partnership with GENEWIZ as combining our offers brings a unique experience to researchers. After submitting their samples for sequencing to GENEWIZ, customers will be able to interact directly with their results and explore the biology of their experiments, in addition to receiving raw data files.
By working together, GENEWIZ and ROSALIND are providing a totally new and fast approach that removes many difficulties with 10x Genomics single-cell assays, allowing researchers to focus on the biology. To me, going back to your first question, this is exactly why I was excited to join the team and develop the platform. Biologists need support to take full advantage of all the great genomics tools available today without having to worry about all the technical steps required to obtain useful results. We need to come up with easy solutions to go from sample to interpretation, and we’re providing exactly that with the GENEWIZ-ROSALIND partnership.
Learn more about GENEWIZ’s scRNA-Seq service leveraging ROSALIND’s data analysis platform to quickly and easily analyze your scRNA-Seq data.
Daily Insight
Recent Posts
Recent Post Tags
- next generation sequencing
- gene synthesis
- rna sequencing
- sanger sequencing
- transcriptomics
- aav-itr
- itr sequencing
- proteomics
- single-cell sequencing
- aav plasmid prep
- antibody discovery
- cell and gene therapy
- olink
- quantitative pcr (qPCR)
- rna therapeutics
- PacBio
- circular rna (circRNA)
- digital pcr (dPCR)
- lentivirus
- long-read sequencing
- mRNA
- oxford nanopore sequencing (ont)
- pcr + sanger sequencing
- whole plasmid sequencing