A Beginner’s Guide to Artificial DNA Synthesis

Basics of Synthetic DNA and Gene Synthesis
Artificial DNA synthesis, a fundamental tool of synthetic biology, enables scientists to create DNA molecules of virtually any sequence without a template. Construction begins with the base-by-base synthesis of oligonucleotides (oligos), followed by assembly into double-stranded DNA (dsDNA) fragments. These custom DNA fragments can be used directly, cloned into vectors, or assembled into larger constructs to serve a variety of research uses (see table below).
| Type of Synthetic DNA | Structure | Research Uses |
|---|---|---|
| Oligonucleotides | Single-stranded, <150 nt | Primers for PCR or sequencing Cloned into vectors for oligo pools qPCR probes Assembly into DNA fragments |
| DNA fragments | Double-stranded, linear | Donor template for gene editing Template for in vitro transcription qPCR standards Assembly of larger DNA fragments Cloned into vectors for gene expression |
| Cloned gene products | Double-stranded, circular | Recombinant protein production Transgenic gene expression gRNA constructs for CRISPR editing |
Compared to classical molecular biology techniques like site-directed mutagenesis, gene synthesis technology generates novel sequences with an unmatched combination of speed, precision, and flexibility. Below, we’ll explore how linear and cloned DNA fragments are made and discuss what factors to consider favoring project success.
DNA Synthesis Process
A gene synthesis project has both wet and dry lab components. It can be broken down into the following steps:
- Define project goals
- Design the sequence
- Synthesize oligos
- Assemble oligos into linear fragments and larger constructs (if applicable)
- Verify the sequence of the gene fragment or cloned product
1. Define Project Goals
Key Considerations
- Communication with collaborators
- Project timeline planning
No matter your molecular target, properly scoping your project will promote success by minimizing revisions. Be sure to consider factors like timelines for downstream assay work or collaborator availability in addition to the overarching project deadlines. It may be more efficient to prioritize the gene products in a specific synthetic order or use a combination of synthetic and molecular techniques to construct them. Speed is normally critical for a gene synthesis project, but a high-quality final product is just as important to reduce repeat syntheses or failed experiments.
2. Sequence Design
Key Considerations
- Literature knowledge and precedence
- Sequence and codon optimization
Once you understand the full scope of your project, it’s time to map a route to synthetic success. A survey of the literature will reveal known issues in the synthesis of your target sequence and how to choose the best codons for optimized expression in your target organism. After a literature survey, further analyze the coding DNA sequence (CDS) for critical elements such as repeats, guanine-cytosine (GC) nucleotide content, unstable motifs or secondary structures, and unintended restriction enzyme sites that will complicate cloning. Access to a free codon optimization tool allows you to skip the literature review. It automatically improves the sequence for optimal synthesis and downstream performance in your experiments. Once optimized, the full nucleotide sequence is then deconstructed into a series of overlapping oligo sequences.
3. Oligo Synthesis
Key Considerations
- Access to instrumentation
- Planning a synthetic route
With a route planned, it’s time to roll your sleeves up and get to synthesizing. Oligo synthesis is a highly automated process. The oligo chain is built on a solid support through cycles of chemical reactions involving precursors called phosphoramidites (Figure 1). With current technologies, it’s not possible to achieve 100% efficiency in each cycle of reactions, leading to a subpopulation of oligos with truncations and point mutations. This issue worsens with oligo length since more cycles are needed to build the molecule. As a result, there is a tradeoff between using longer oligos, which generally simplifies assembly, and the sequence fidelity of the final product.
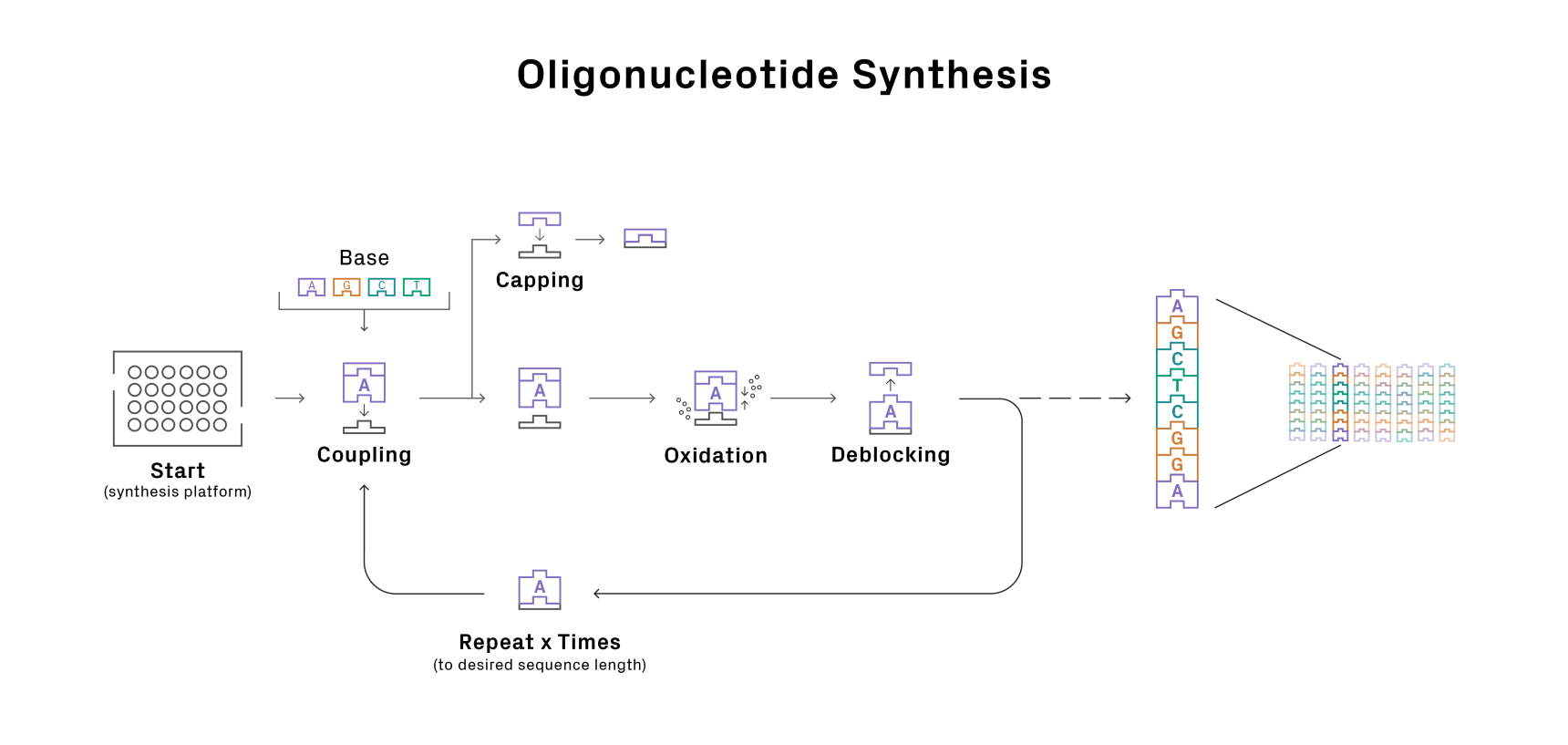
Figure 1: Oligonucleotide synthesis cycle.
4. Gene Assembly
Key Considerations
- Linear DNA fragments vs. cloned DNA
- Experience with and access to plasmid vectors
Due to technical limitations of phosphoramidite-based oligo synthesis, it’s best to build dsDNA fragments (>150 nt) via polymerase cycling assembly. Purified oligos are mixed and allowed to anneal to one another. Primer extension with DNA polymerase generates dsDNA, which is then amplified by PCR. Depending on the application, the linear products can be used “as-is,”, used as building blocks for larger fragments via methods like Gibson assembly, or cloned into a vector. Since gene fragments are a heterogeneous mixture of DNA molecules, it’s generally preferable to clone them prior to downstream use.
5. Sequence Verification
Key Considerations
- Access to instrumentation
- Rigor of quality control based on product
Lastly, the final synthetic DNA product should be analyzed for sequence accuracy via Sanger sequencing or next-generation sequencing. The latter can also quantitate the diversity of a library of DNA products. Restriction enzyme digests are a good complement to sequencing, especially when the vector backbone is not fully sequenced.
Synthetic DNA Services
The steps above can be laborious to perform in-house. Instead, to save time and focus on the downstream applications of synthetic products, consider a service provider for gene synthesis. This provider should be considered a partner in your research, and as such, they should help you achieve the project goals of today and beyond. To enable your success, think about access to local support, reputation for quality and reliability, expertise in synthesis and cloning, as well as the ability to support long-term and multi-institution projects that have goals in pre-clinical and clinical trials with CLIA or GLP requirements.
Key Takeaways
- Artificial DNA synthesis has the power to generate virtually any DNA sequence
- Planning, optimized design, and rigorous sequence verification are critical to efficient gene synthesis and project success
- A service provider can dramatically reduce error, labor, and time required for synthesizing genes as compared to do-it-yourself approaches
See how codon optimization can improve your next synthetic DNA project by downloading our technical note on enhancing protein expression.
Daily Insight
Recent Posts
Recent Post Tags
- next generation sequencing
- gene synthesis
- rna sequencing
- sanger sequencing
- transcriptomics
- aav-itr
- itr sequencing
- proteomics
- single-cell sequencing
- aav plasmid prep
- antibody discovery
- cell and gene therapy
- olink
- quantitative pcr (qPCR)
- rna therapeutics
- PacBio
- circular rna (circRNA)
- digital pcr (dPCR)
- lentivirus
- long-read sequencing
- mRNA
- oxford nanopore sequencing (ont)
- pcr + sanger sequencing
- whole plasmid sequencing